10 Best Free Phylogenetic Tree Viewer Software For Windows
Here is a list of Best Free Phylogenetic Tree Viewer Software For Windows. Using these software, you can view, analyze, and modify the phylogenetic trees of different species. By analyzing the evolutionary trees of different species, you can understand the process of evolution that took place.
If I talk about the supported formats, these software support multiple phylogenetic tree formats. Some of these formats include NEXUS, Phylip, Dendro, Text, NeXML, and many more. After opening a phylogenetic tree, you can view it in multiple layouts. Some of these layouts include Equal Angle Tree, Average Consensus, Balanced Confidence Network, Rectangular Cladogram, Slanted Cladogram, Circular Phylogram, etc.
Some of these software feature a Magnifying Glass. Move this magnifying glass over the tree to see the magnified view of that region. Besides this, some software come with Find and Replace feature. This feature lets you find a text in the tree and replace it with some other text.
You can save the modified tree in the formats supported by these software or export it in different image formats. These image formats include BMP, GIF, JPEG, PNG, etc. One of these software also lets you export the selected parts of the tree, like Taxa, Sets, Splits, Network, etc.
My favorite Phylogenetic Tree Viewer Software:
TreeView is my favorite phylogenetic tree viewer software. It not only lets you view a phylogenetic tree, but also lets you create a phylogenetic tree on your own. Moreover, it supports multiple input formats including NEXUS files, PHYLIP files, Text files, etc. Besides this, you can also save the tree as a graphic in EMF or WMF format.
You may also like some best free Genetic Heredity Calculator, Punnett Square Calculator, and Medical Manager software for Windows.
TreeView
TreeView is a free phylogenetic tree viewer software for Windows. In this software, you can open and edit the evolutionary trees of different species. Hence, by analyzing the evolutionary trees, you can study how the process of evolution has taken place in different species. It opens every phylogenetic tree in a new window which lets you analyze and edit multiple evolutionary trees without closing any of them.
TreeView is not only a phylogenetic tree viewer, but also a phylogenetic tree maker software.
Steps to create a phylogenetic tree from scratch are as follows:
In order to create a phylogenetic tree, you first have to create a text file containing a list of names of all the species. The names may be biological names or common names. You can use Wordpad, Notepad, Notepad++, or any other software for this purpose. All the names should be written in such a manner that one row contains only one name. After creating a list, save it in TXT format. Now, launch the TreeView and go to File > Import > List of Taxon names and select the text file which you have created and click on Open button. After opening the file, you will see that all the names of species are connected to a single node with separate branches. Now, go to Edit > Edit Tree to correct the phylogenetic tree by joining different branches. Multiple tools are provided on the toolbar for editing an evolutionary tree. These include Move branch, Collapse branch, Collapse clade, Reroot tree, Rotate descendants, Swap descendants, etc. A Text tool is also provided to write a text at different nodes of the tree. After finishing editing, save it from the File menu.
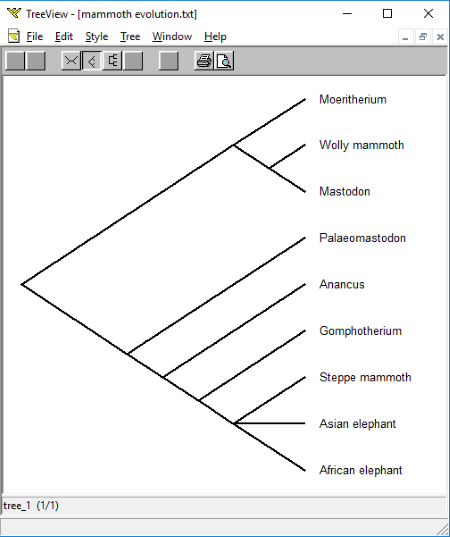
See the above screenshot in which I have tried to create an evolutionary tree of Mammoth.
Four different viewing options are available in the software, namely, Unrooted, Slanted Cladogram, Rectangular Cladogram, and Phylogram.
General features of this free cladogram maker software:
- It supports multiple formats: NEXUS files (,tre, .trees), PHYLIP files (.phy), CLUSTAL W (.dnd, .ph, .phb), Henning86 (.hng), and Text files (.txt).
- You can change the font size and font style.
- It lets you copy the evolutionary tree simply by pressing the Ctrl+C key.
- Print and Print Preview options are also available in the software.
- You can also save the tree as a graphic in EMF and WMF formats.
Unipro UGENE
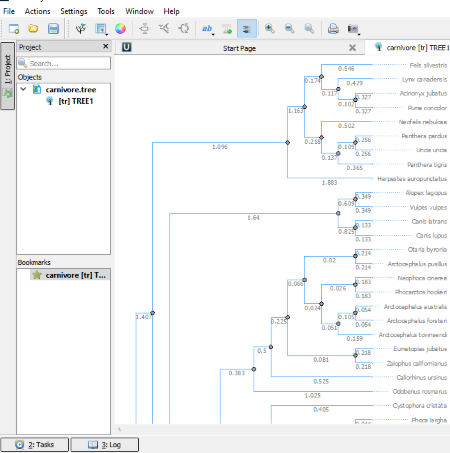
Unipro UGENE is a featured phylogenetic tree viewer software which is intended to do multiple tasks. It is not only a phylogenetic tree viewer software, but also a DNA sequence analyzer plus creator and workflow creator software.
Since it is a multipurpose software, it supports multiple file formats. Over 30 file formats are supported by Unipro UGENE. Some of these include NEXUS, Newick standard, PHYLIP interleaved, PHYLIP Sequential, Plain text, Position frequency matrix, UGENE database, Vector NTI, pDRAW, GFF, GTF, BAM, FASTA, FASTQ, GenBank, MSF, Mega, etc.
It comes with three types of tree layouts: rectangular phylogram/cladogram, circular phylogram/cladogram, and unrooted phylogram/cladogram. You can zoom in or out the tree by using the scroll wheel of the mouse. In the branch settings, you can change the color of branches, width of the tree and height of the tree. Moreover, the color, font, and style of the text can also be changed. It also displays the length/distance of each branch. You can select whether to show or hide the names of species and distances of branches.
Screen Capture: You can capture the screen in various formats, like PNG, BMP, JPG, PDF, TIFF, SVG, etc. The entire tree capturing option is also available, but this can save the tree only in SVG format.
You can save and export the project in its own supported format. Besides this, you can also print the tree.
Unipro UGENE is a free and open source phylogenetic tree viewer software.
SplitsTree4
SplitsTree4 is another free phylogenetic tree viewer software for Windows. It lets you view, modify, and analyze the evolutionary trees in multiple formats. These formats include FastA files (.fa, .fasta), Clustal Alignment Files (.aln), Phylip Sequences Distance Matrix files (.dist, .dst), MrBayes Partition files (.parts), Newick Tree files (.new, .tre, .tree), Nexus files (.nxs, .nex, .nexus), Reticulate Network files (.rnet), Single line sequences files (.txt), etc.
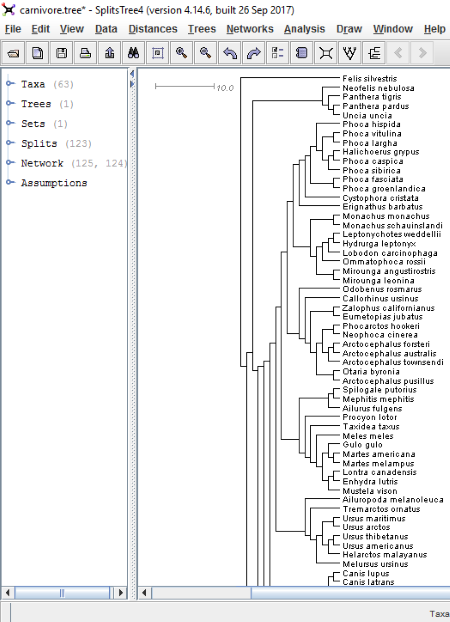
The phylogenetic tree diagram/phylogenetic tree network can be displayed in eight different split methods, namely, Cluster Network, Convex Hull, Equal Angle, Hybridization Network, No Graph, Phylogram, Reticulate Network, and Rooted Equal Angle. By default, the diagram is displayed in Equal Angle split tree method. In order to change it, go to Draw menu > Select Trees and click on Splits tab. There you can see a drop-down menu from where the splits transformation method can be selected. After selecting the type of split, click on Apply button. You can also modify each of these split tree methods by changing their properties, like percentage of offset, maximum reticulations per tangle, layout, maximum angle, out-group, etc. Do note that each of these mentioned properties is not applicable to all of the split transformation methods.
Apart from the splits transformation method, multiple network types are also available for phylogenetic tree diagrams. Some of these include Equal Angle Tree, Average Consensus, Balanced Confidence Network, Consensus Network, Consensus Tree, Filtered Super Network, etc. Simply go to Draw > Select Trees and click on the Method tab in the second row in the window thus opened.
On the left panel of the software, you can easily view the number of Taxa, number of splits, number of networks, etc. in a tree. Expand these to view more details.
Some useful features of SplitsTree4:
- Magnifier: This feature is available in the View menu. On enabling this feature, a magnifying glass becomes available on the interface. Move this glass to the different regions of a tree to see the magnified view of that region. Click on the above screenshot to see how this magnifying glass looks like.
- Filter Taxa: Go to Data > Filter Taxa and select the taxa which you want to hide from the phylogenetic tree.
- Edit Traits: This option is available in the Data menu. You can add the new traits or remove the existing ones.
- Find and Replace: With the help of this feature, you can find a text in the tree and replace it with any other text. You can make your search case sensitive, whole words only, and regular expression.
You can save the modified phylogenetic tree in its own format. Besides this, export to image feature is also provided. You can export the tree in EPS, GIF, JPEG, PNG, BMP, PDF, and SVG formats. Moreover, you can also export the selected parts of the tree such as Taxa, Sets, Splits, Network, etc.
Dendroscope
Dendroscope is yet another free phylogenetic tree viewer software for Windows. It supports the following tree formats: Dendro (.dendro), Nexus (.nexus), Newik (.newick), NeXML (.nexml), and Text (.txt).
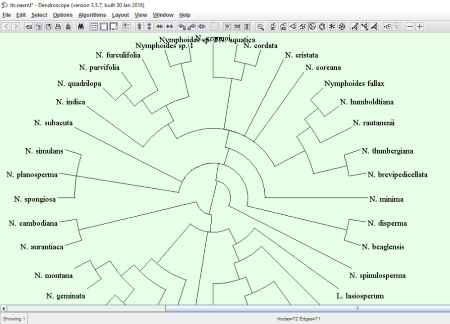
Dendroscope comes with some example evolutionary trees, which you can view and analyze. These example files include fishes, grasses, and nymphoides. You can also modify the type of layout of a phylogenetic tree. As compared to other phylogenetic tree viewer software in this list, this one features a good number of tree layouts. These include Rectangular Phylogram, Rectangular Cladogram, Slanted Cladogram, Circular Phylogram, Circular Cladogram, Inner Circular Cladogram, Radial Phylogram, and Radial Cladogram. All these layouts are available in the Layout menu and the toolbar of the software. Besides this, you can also align the entire network to left, right, and in random order. Talking about the network layout, three types of algorithms are given, namely, 2008, 2009, and 2010 algorithms.
Basic features of this free phylogenetic tree analysis software:
- You can copy the tree to the clipboard.
- On moving the Magnifying Glass on the particular area of the tree, the software displays the magnified view of that region.
- Find and Replace option is also provided in the software, so that you can find a particular name in the tree and replace it with another text. Following types of search options are given: case sensitive, whole words only, and regular expression.
- You can save the modified evolutionary tree in any of the above-mentioned tree formats.
- It also lets you print the cladogram.
- You can also export the phylogenetic tree in some popular image formats, like BMP, GIF, JPEG, PNG, etc. Besides this, you can also export the tree to PDF.
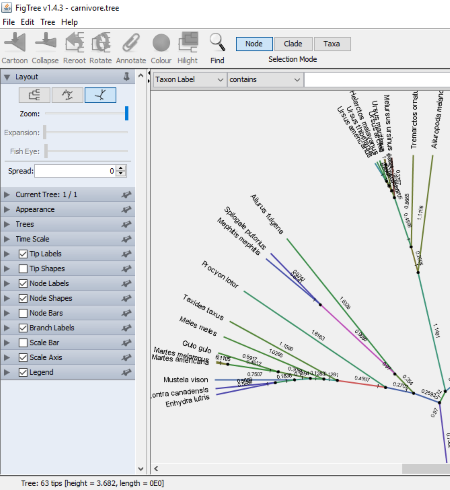
FigTree
FigTree is a JAVA based application to view phylogenetic trees. The number of formats supported by the software is not mentioned. I tried .tree format and that was opened in the software. You can analyze multiple trees simultaneously in this software, as it opens each new tree in a new window.
Phylogenetic Tree Layout: Three types of layouts are available in the software, namely, Rectangular tree layout, Polar tree layout, and Radial tree layout. All these layouts are accessible on the left panel of the software. The view of each of these layouts can be varied by changing their properties. These properties include Expansion, Fish Eye, Root Length, Curvature, etc. You will find different properties available for different layouts.
In the Appearance section, you can change some general factors of the phylogenetic tree such as line width, background colors, etc. Besides this, you can also highlight different regions of the tree and the names of different species with different colors.
Annotate is a useful feature of FigTree. Using this feature, you can change any of the names in the tree. Simply select the name which you want to change and click on Annotate button on the toolbar and replace it with another name. You can also import the annotations into the software, but input format for this purpose is not mentioned.
By using the Find option, you can search for the following: Texon Label, Branch Length, Node Age, Name, Height, Any Annotation, etc. If you want, you can perform Case Sensitive searches. Apart from this, some filters are also available such as contains, starts with, end with, regular expression, etc.
You can export the phylogenetic tree in PDF, SVG, PNG, and JPEG formats.
BayesTrees
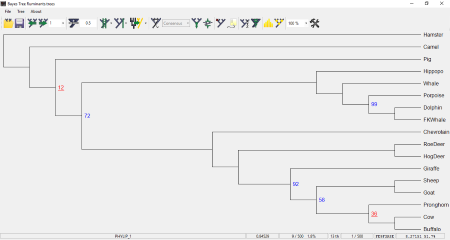
BayesTrees is a very simple phylogenetic tree viewer software for Windows. You can open a phylogenetic tree in this software with any of the following formats: Nexus Trees and Bayes Trees. It comes with some example phylogenetic trees which you can view in this software and analyze.
Phylogenetic Tree Style: It features 6 different tree styles, namely, Normal, Arc, Curved, Sagging, Balloons, and Slanted. All these tree styles are available in a drop-down box on the toolbar menu.
You can also ladderise the tree to any of the left or right directions.
The modified tree can be saved in the following formats: Nexus trees, Bayes trees, Strict Phylip, and Relaxed Phylip. Besides this, you can also save the tree in PNG, BMP, JPG, TIFF, GIF, EMF, etc. image formats.
TreeGraph 2
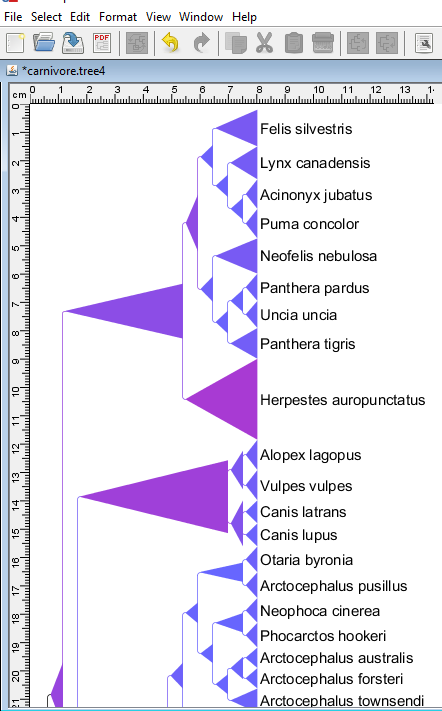
TreeGraph 2 is one more free phylogenetic tree viewer software for Windows which lets you view and edit the phylogenetic trees. It supports various formats including TreeGraph 2 XML, PhyloXML, NeXML, Nexus File, and Newick file formats.
You can edit the tree by applying different changes. Some of these are given below:
- You can add different colors to different branches or color the entire tree with a single color. Coloring option is available in the Format menu.
- You can also set the minimum and maximum distance values by selecting different node/branch data. For example, branch line widths, minimal branch lengths, minimal space above branches, node line widths, corner radius of nodes, leaf margin (left, right, top, and bottom), etc. The distance values can be set in millimeters only.
- Two types of view are available in this freeware, namely, Rectangular Cladogram and Phylogram/Cladogram.
- You can delete the existing taxa or add new taxa to the tree.
- Search option is also available. Using this feature, you can search the name of any species in the tree. You can also apply the following filters to the search: case sensitive, words only, and include internal node names.
After editing the graph, you can save it in TreeGraph XML format or export it in PDF, Nexus file, and Newick file formats.
MultiDendrograms
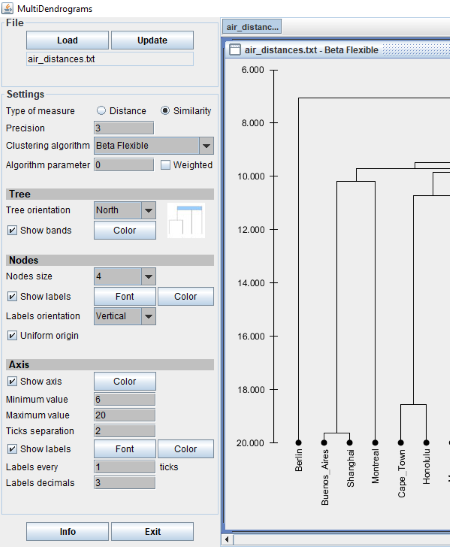
MultiDendrograms is the next free phylogenetic tree viewer software for Windows. It comes with some pre-loaded example files. You can open and analyze them. To open a file in the software, click on LOAD button provided. The example files are in TXT formats that means it supports the files in TXT format. But this is not the case, when I tried to open other TXT files in the software, it shows an error message.
After opening a phylogenetic tree, you can change the tree orientation in any of the four directions (East, West, North, and South), change the view of tree by selecting any of the clustering algorithms (versatile linkage, single linkage, complete linkage, geometric linkage, centroid, etc.), vary the nodes’ size, show/hide labels, etc. It does not have any auto-refresh option. Hence, you have to click on Update button after any change you make.
Save: You can save the phylogenetic tree in TXT, Newick tree, JSON tree, PNG, JPG, and EPS formats.
T-REX
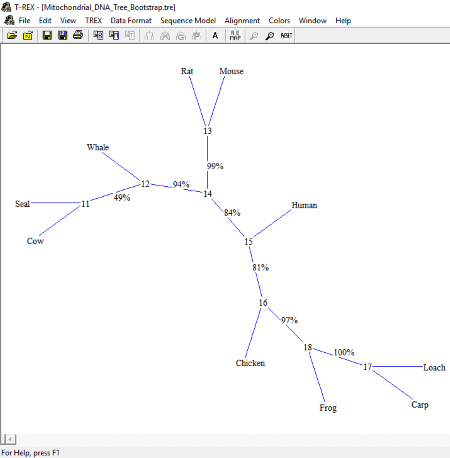
T-REX is another free software to view phylogenetic trees. It supports Newick tree and T-Rex (.tre) file formats. It has various sample files of phylogenetic trees. You can open them in the software and analyze them.
Talking about the tree style, four types of tree styles are available: Hierarchical Vertical, Hierarchical Horizontal, Axial, and Radial. These tree styles are available in Edit menu > Redraw the Map.
You can also change the color of names of species and branches of the tree.
The opened tree can be saved in Newick format or in its own T-REX (.tre) format. Besides this, you can also take a printout of the tree.
Treefinder

Treefinder is yet another free phylogenetic tree viewer software. Formats supported by this software are not mentioned, but I tried Tree and TXT formats and both of these formats are supported by it.
Steps to open a phylogenetic tree in Treefinder is different from other phylogenetic tree viewer software in this list. Go to File > Open Image and select the supported file format. The phylogenetic tree is then opened as an image. Since the phylogenetic tree is opened as an image, you cannot modify the tree. But, you can change the style of the tree. This can be done by changing the topology of the phylogenetic tree. Various topologies include Midroot, Reroot, Swap, Collapse, etc. You will find these topologies in Redraw option of the View menu.
It lets you export the tree in different formats which include TL Tree List, PHYLIP, Newick, NEXUS, etc.
About Us
We are the team behind some of the most popular tech blogs, like: I LoveFree Software and Windows 8 Freeware.
More About UsArchives
- May 2024
- April 2024
- March 2024
- February 2024
- January 2024
- December 2023
- November 2023
- October 2023
- September 2023
- August 2023
- July 2023
- June 2023
- May 2023
- April 2023
- March 2023
- February 2023
- January 2023
- December 2022
- November 2022
- October 2022
- September 2022
- August 2022
- July 2022
- June 2022
- May 2022
- April 2022
- March 2022
- February 2022
- January 2022
- December 2021
- November 2021
- October 2021
- September 2021
- August 2021
- July 2021
- June 2021
- May 2021
- April 2021
- March 2021
- February 2021
- January 2021
- December 2020
- November 2020
- October 2020
- September 2020
- August 2020
- July 2020
- June 2020
- May 2020
- April 2020
- March 2020
- February 2020
- January 2020
- December 2019
- November 2019
- October 2019
- September 2019
- August 2019
- July 2019
- June 2019
- May 2019
- April 2019
- March 2019
- February 2019
- January 2019
- December 2018
- November 2018
- October 2018
- September 2018
- August 2018
- July 2018
- June 2018
- May 2018
- April 2018
- March 2018
- February 2018
- January 2018
- December 2017
- November 2017
- October 2017
- September 2017
- August 2017
- July 2017
- June 2017
- May 2017
- April 2017
- March 2017
- February 2017
- January 2017
- December 2016
- November 2016
- October 2016
- September 2016
- August 2016
- July 2016
- June 2016
- May 2016
- April 2016
- March 2016
- February 2016
- January 2016
- December 2015
- November 2015
- October 2015
- September 2015
- August 2015
- July 2015
- June 2015
- May 2015
- April 2015
- March 2015
- February 2015
- January 2015
- December 2014
- November 2014
- October 2014
- September 2014
- August 2014
- July 2014
- June 2014
- May 2014
- April 2014
- March 2014